Introduction¶
This is a full example strategy showcasing flexible gating of a sample along lymphocyte and T-lymphocyte lineages with both advanced and simpler features.
Some light commentary has been added to the various jupyter notebook cells.
Most pattern recognition approaches here uses some default and sanity limits through if statements, these are typically based on FMOs, or based on experience from iterative attempts of gate random selections of samples.
[1]:
import aligater as ag
AliGater started in Jupyter mode
The code block below contains a set of gating steps that has been broken out from the rest of the strategy into their own functions, this might be useful to compartmentalize specific problems. However, it can look a bit confusing.
You can skip looking at this cell now and return to it when these functions are being called later.
[2]:
# This function simply creates a list of [x,y] coordinates from a given [xstart,ystart] to
# [xend, yend] with even spacing between
#Such lists can be supplied to gatePointList
def create_vPL(startx, endx, starty, endy, bins = 300):
x_coord = []
y_coord = []
xstep = (endx-startx)/bins
ystep = (endy-starty)/bins
for i in ag.np.arange(startx,endx,xstep):
x_coord.append(i)
for i in ag.np.arange(starty,endy,ystep):
y_coord.append(i)
assert len(x_coord) == len(y_coord)
vPL = ag.np.column_stack([x_coord,y_coord])
return vPL
# This function contains broken-out pattern recognition methods to solve a specific gate that's repeated multiple times
def CD4gate(fcs, gate, manual_xlim = None, manual_ylim_left = None, manual_ylim_right = None, fileName = None):
xlim = ag.valleySeek(fcs,"CD45RO",gate,interval=[500,2000],scale='logish')
ylim = ag.valleySeek(fcs, "CD45RA",gate,interval=[500,1000],scale='logish') #Change 2000 -> 1000 from phase I > phase II
solution = ag.variableQuadGate(fcs, ['','','',''],"CD45RO",CD45RA,parentGate=gate,scale='logish',threshList=[xlim,xlim,ylim,ylim],testRange=[0,1000],position='left',only_solution=True)
testUpperLim = 1500
testLowLim = 500
naiveCD4, tmp1, memoryCD4, tmp2, solution = ag.variableQuadGate(fcs,['naiveCD4', 'tmp1', 'memoryCD4', 'tmp2'],"CD45RO",CD45RA,parentGate=gate,scale='logish',threshList=solution,testRange=[testLowLim,testUpperLim],position='right',only_solution=False, filePlot=fileName)
while len(memoryCD4()) < 0.4*len(tmp1()):
solution[3] += 500
testUpperLim = solution[3]+1000
testLowLim = solution[3]
naiveCD4, tmp1, memoryCD4, tmp2, solution = ag.variableQuadGate(fcs,['naiveCD4', 'tmp1', 'memoryCD4', 'tmp2'],"CD45RO",CD45RA,parentGate=gate,scale='logish',threshList=solution,testRange=[testLowLim,testUpperLim],position='right',only_solution=False, filePlot=fileName)
if manual_xlim is not None:
#In order; bottom x threshold, top x threshold, left y threshold, right y threshold.
solution[0] = solution[1] = manual_xlim
if ylim_left is not None:
solution[2] = manual_ylim_left
if ylim_right is not None:
solution[3] = manual_ylim_right
if any([x is not None for x in [manual_xlim, manual_ylim_left, manual_ylim_right]]):
threshList = solution[0:4]
naiveCD4, tmp1, memoryCD4, tmp2 = ag.customQuadGate(fcs,names=['naiveCD4', 'tmp1', 'memoryCD4', 'tmp2'],threshList=threshList ,xCol="CD45RO",yCol=CD45RA,parentGate=gate, scale='logish', filePlot=fileName)
return naiveCD4, memoryCD4
# This gate is instead very simple, but used many times, so it has been defined as a function to allow easier calls
def FMOCD39(fcs,gate):
output = ag.gateThreshold(fcs, xCol = CD39, yCol = "FSC 488/10-H", name = "CD39FMO", parentGate = gate,
orientation = 'vertical', scale = 'logish', thresh = 700, population = 'upper',
update = False, IgnoreCellLimit = False)
return output
[3]:
# Defining some comfy marker aliases
# This allows to use these shorthand variables instead of the whole string when calling functions
SSC = "SSC 488/10-A"
FSC = "FSC 488/10-A"
CD39 = "BB515 CD39-A"
CD25 = "PE-Cy7 CD25-A"
CD127 = "PE CD127-A"
CCR6 = "PE-Dazzle 594 CCR6-A"
HLADR = "BV650 HLA-DR-A"
CCR7 = "BV711 CCR7-A"
CXCR5 = "BV786 CXCR5-A"
CXCR3 = "BV421 CXCR3-A"
CD194 = "BV605 CD194-A"
CD4 = "BV510 CD4-A"
CD3 = "Alexa Fluor 700 CD3-A"
CD8 = "APC-H7 CD8-A"
CD45RA = "APC CD45RA-A"
Loading an example .fcs file¶
[7]:
fcs = ag.loadFCS(ag.AGConfig.ag_home + "/tutorial/data/example1.fcs", flourochrome_area_filter = True, compensate = True, metadata = False, return_type = "agsample")
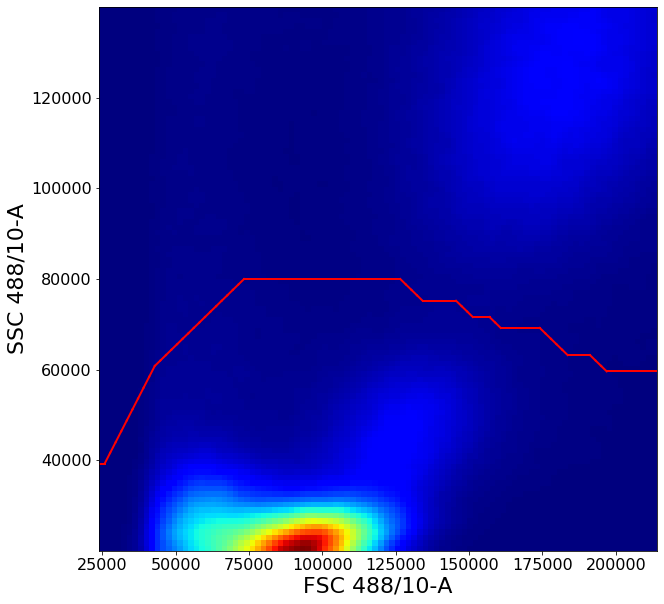
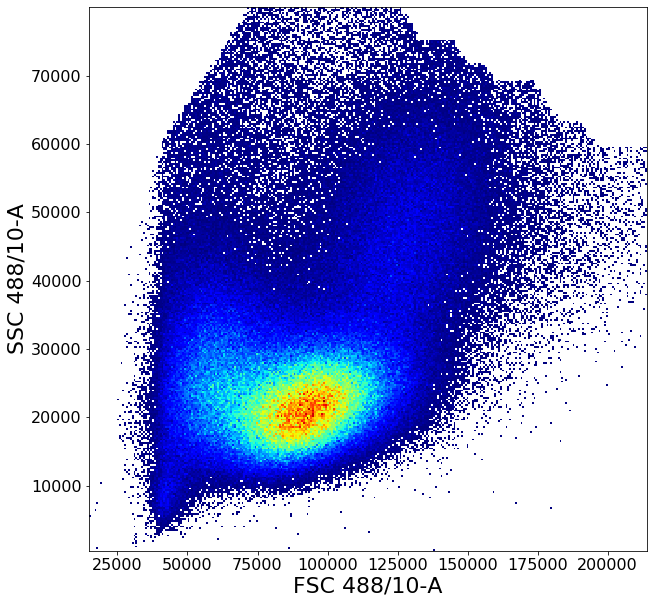
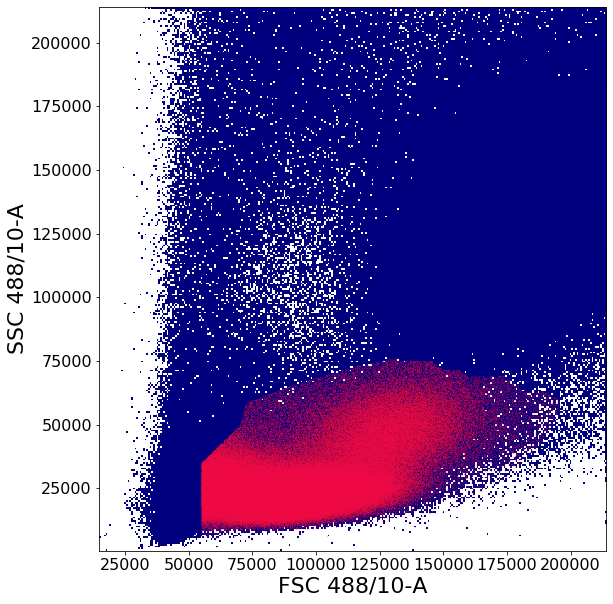
Gating PBMCs using shortest path algorithms.¶
This gate has two purposes;
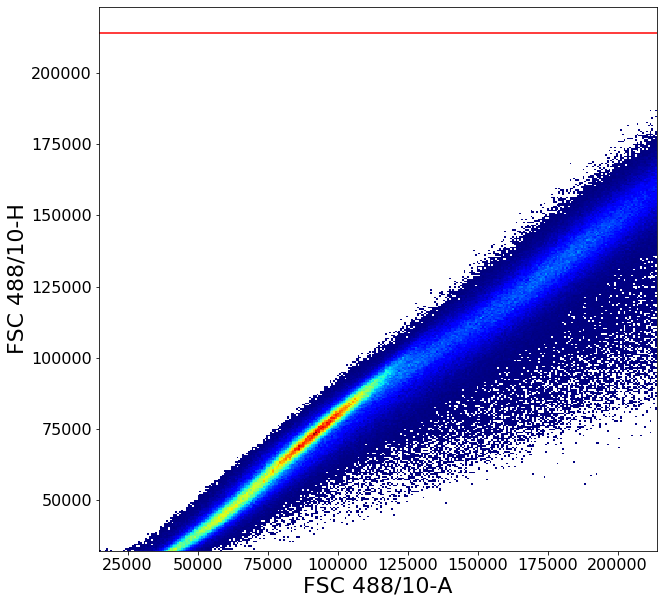
1 - Remove piled up events along the right edge of the plot. Similar to commercial software, AliGater will pile up events higher than some maximum value, which can be configured in the aligater config.
You can always see what this value is set to with below command.
[5]:
ag.AGConfig.ag_maxMeasurement
[5]:
214000
2 - Isolate “Peripheral Blood Mononuclear Cells” (PBMCs). To do this we need to get rid off the high FSC-H intensity cells which are typically neutrophils, then get a reasonably clear circling of the remaining cluster to remove low FSC-A FSC-H events, which are probably debris.
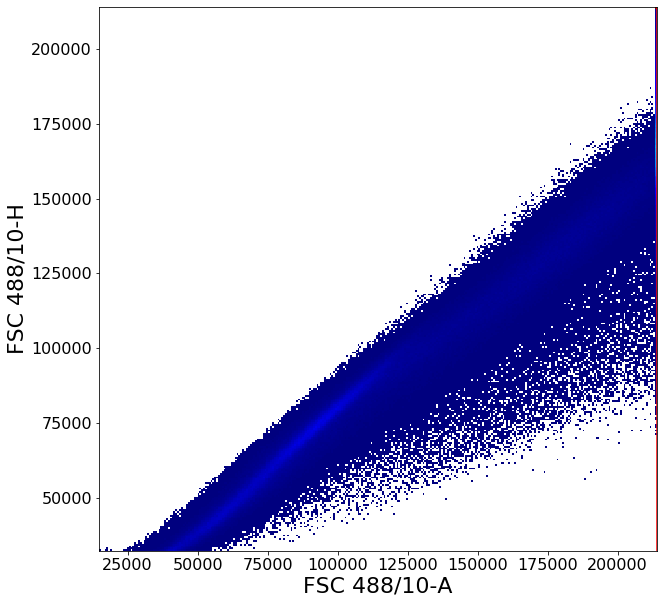
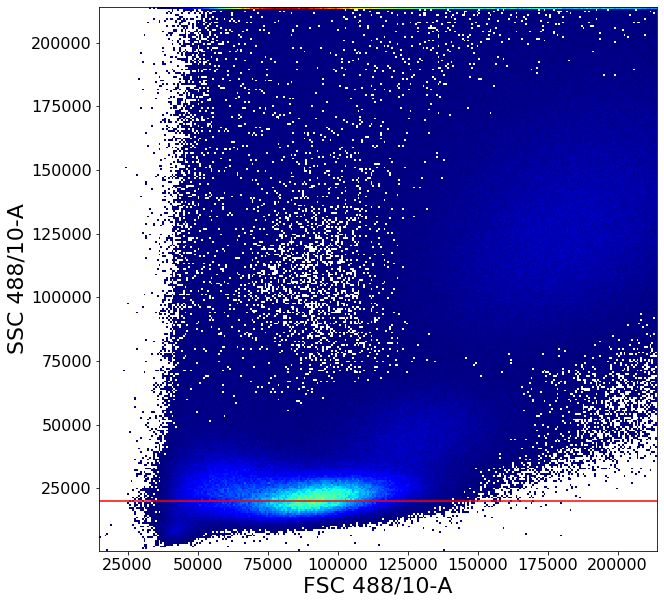
This gate uses some of the more advanced AliGater gates, showcasing HorizontalPath, gatePC, gatePointList as well as gateThreshold and at the end, backGate for visualization purposes.
[6]:
# Treshold gate to remove high SSC-A and FSC-A events
no_clutter1 = ag.gateThreshold(fcs, "no_clutter", xCol = FSC, yCol = SSC, thresh = 214000,
orientation = 'vertical', population = "lower")
no_clutter = ag.gateThreshold(fcs, "no_clutter", xCol = FSC, yCol = SSC, parentGate = no_clutter1,
thresh = 214000, orientation='horizontal', population="lower")
# Horizontal path calculates a Dijstra's shortest path with constraints from startY to endY
# The constraints in this case are:
# 1) Initial 'zoom in' on SSC-A between 20,000 and 140,000
# 2) maxstep 2
# 3) phi 0.1
# 4) direction 'both'
# I.e. The path can move both up and down as it traverses, for each step on the x axis it can take a maximum of +-2 steps in the y direction
# Longer steps are slightly penalized over shorter steps
PBMCstep1 = ag.horizontalPath(fcs, name = "PBMCstep1", xCol = FSC, yCol=SSC, parentGate = no_clutter,
population='lower', startY = 40000, endY = 60000,
xboundaries = None, yboundaries=[20000,140000],
leftRight = True , direction = 'both', maxStep = 2, phi = 0.1,
bins = 100, sigma = 1, scale = 'linear', T = 1000)
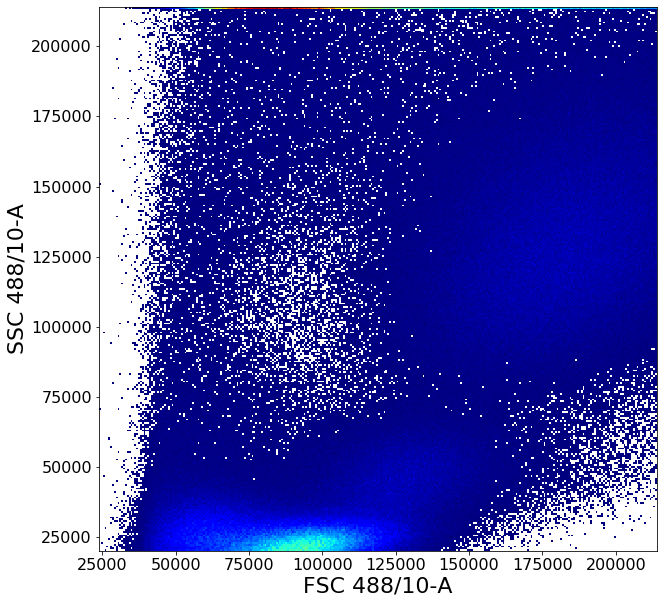
# To remove debris in the bottom-left corner, a custom fixed gate is defined through gatePointList
# First a list, "vPL", of x and y coordinates is defined then supplied to the function
vPL = [[55000,0],[55000,35000]]
vPL2 = create_vPL(55000,70000,35000,50000)
vPL3 = create_vPL(70000,90000,50000,120000)
vPL.extend(vPL2)
vPL.extend(vPL3)
vI = ag.gatePointList(fcs(), xCol = FSC, yCol = SSC, vPL = vPL, population = 'upper', vI = PBMCstep1(), bhorizontal = False)
# gatePointList is a lower lever function and that doesn't return aligater.AGSample objects and
# instead returns a list-like index, which points to events in the sample dataframe that pass the gate
# This is useful to known if custom gates are constructed
# Convert an list-like integer index to the pandas dataframe of the AGSample to an AGGate like this:
PBMCstep2 = ag.AGgate(vI, PBMCstep1, xCol=FSC, yCol=SSC, name="PBMCstep2")
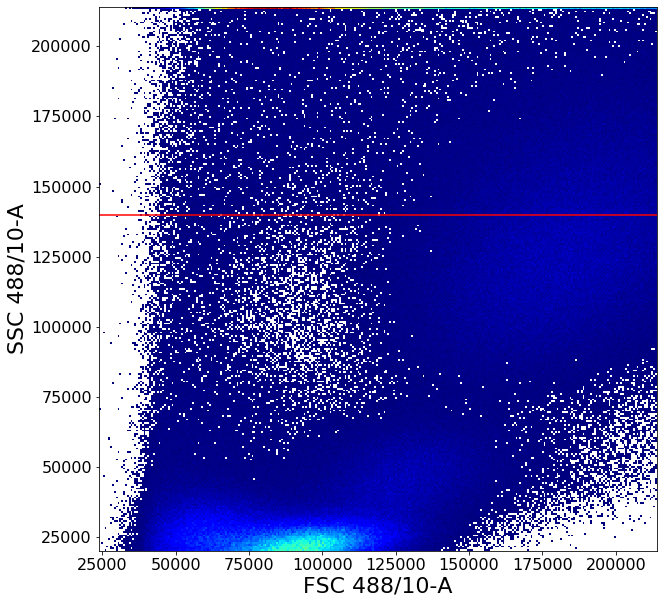
# Finally to further defined the PBMC cluster a principal component based ellipsoid gate is drawn
# This behaves similarly to a 2-D gaussian mixed model with some useful constraints
# In this case the semiaxis of the ellipsoid is scaled to 4 and 3.5 times the length of the first two eigenvectors through the widthScale and heightScale arguments
# The angle of the ellipsoid is also slightly nudged up by 3 degrees through the adjustAngle Argument
PBMC = ag.gatePC(fcs, name="PBMC", xCol = FSC, yCol = SSC, center = 'centroid',
adjustAngle = 3, widthScale = 4, heightScale = 3.5,
parentGate = PBMCstep2)
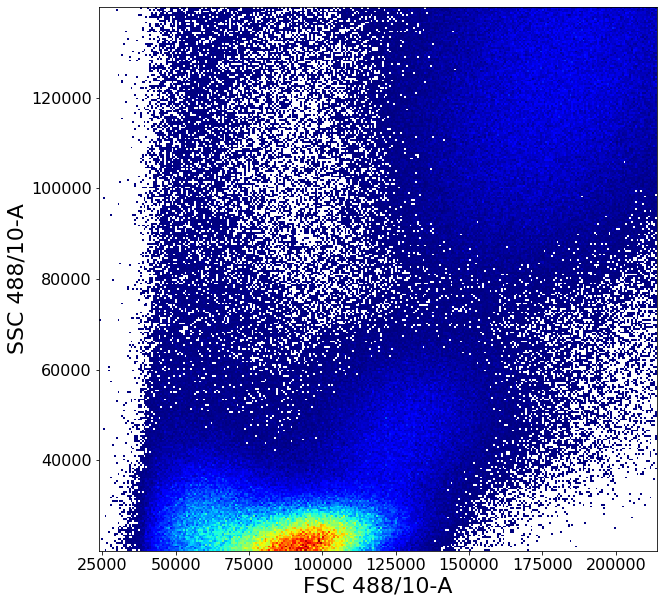
# As an extra visualization step, a backGate call is made to show where the resulting events are on the original plot view
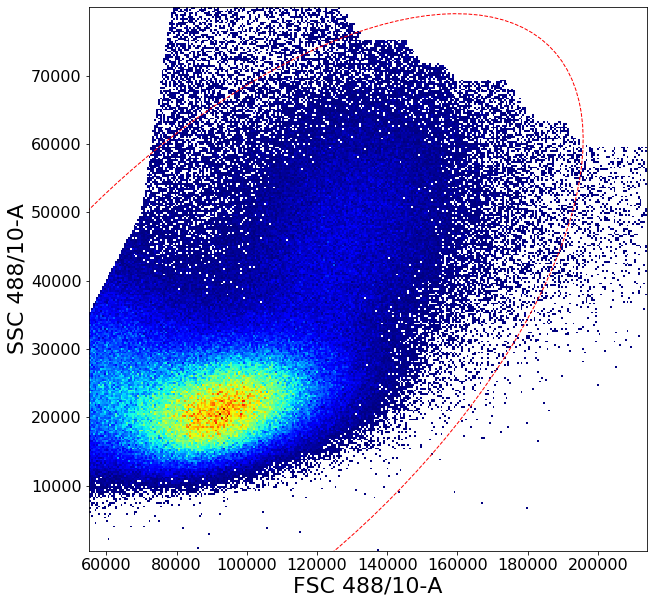
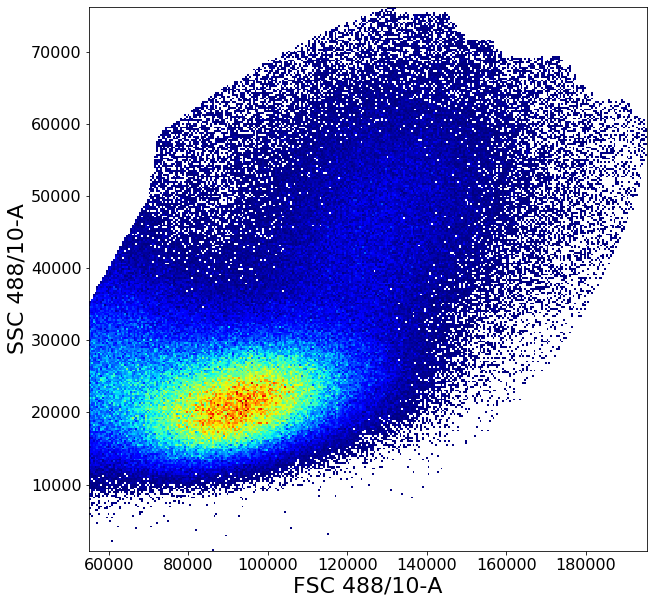
ag.backGate(fcs, population = PBMC, background_population = None, xCol = FSC, yCol = SSC, markersize = 0.1)
<Figure size 720x720 with 0 Axes>
<Figure size 720x720 with 0 Axes>
<Figure size 720x720 with 0 Axes>
<Figure size 720x720 with 0 Axes>
<Figure size 720x720 with 0 Axes>
<Figure size 720x720 with 0 Axes>
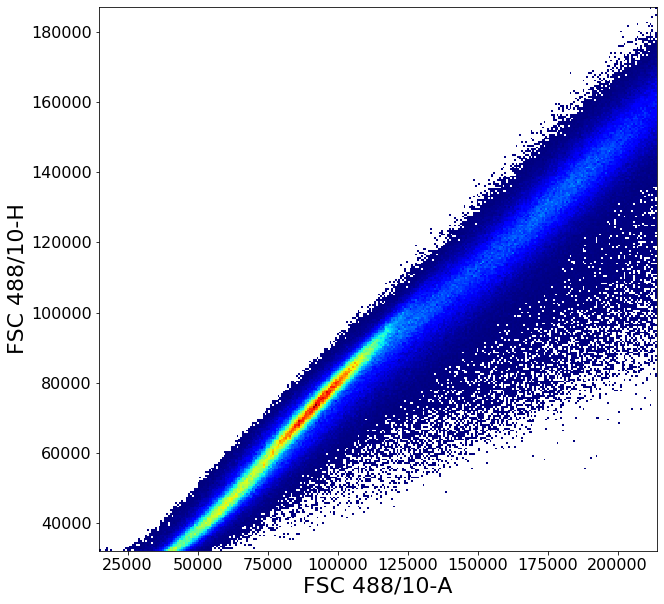
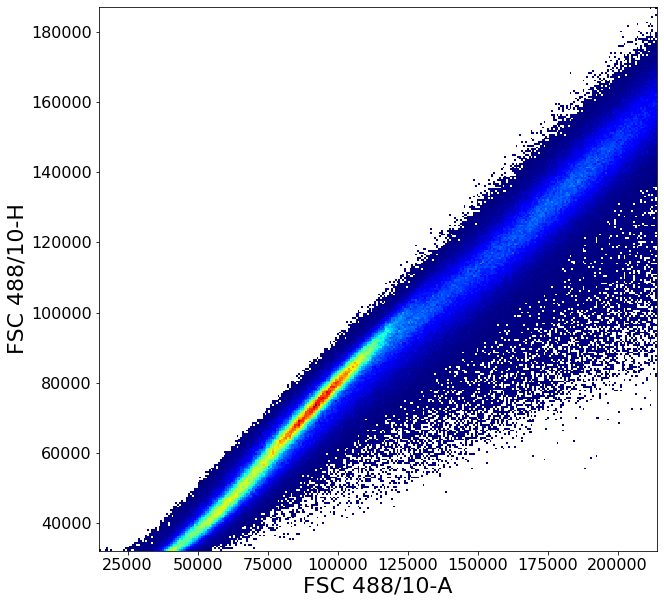
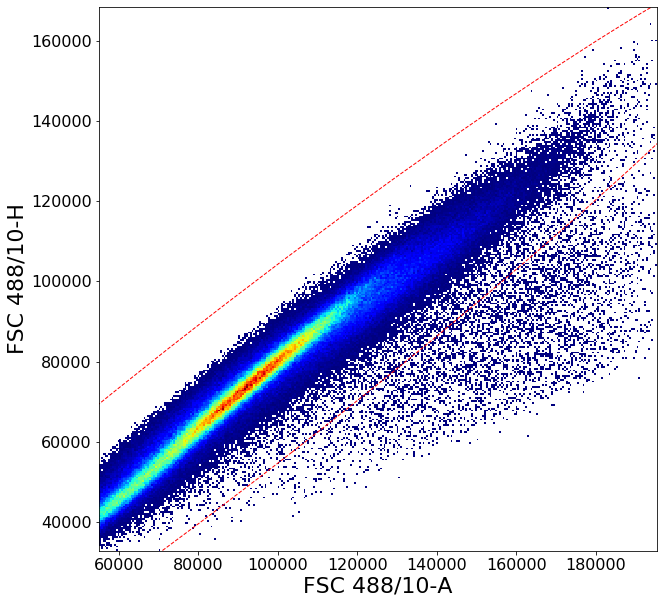
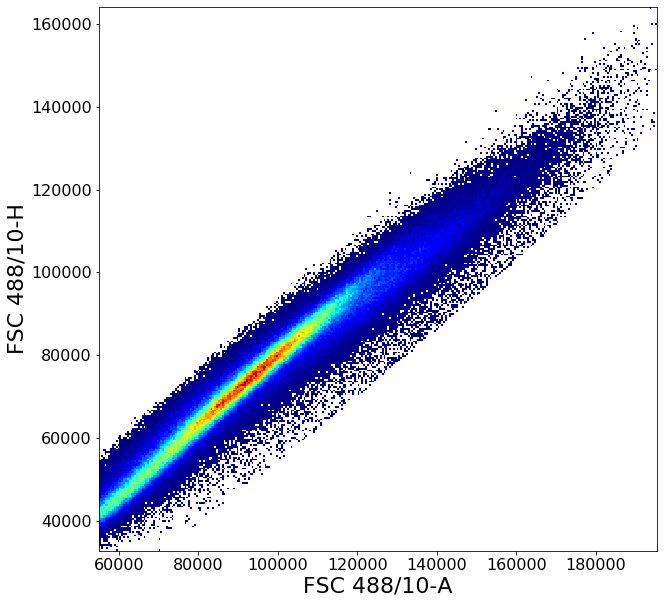
Gating singlets using principal component based clustering¶
Similarly to the gate above, gatePC is used to define singlets from PBMCs
[7]:
singlets = ag.gatePC(fcs, xCol = FSC, yCol = "FSC 488/10-H", name = "singlets", center = 'density',
adjustAngle = 1, widthScale = 6, heightScale = 4.5, parentGate = PBMC)
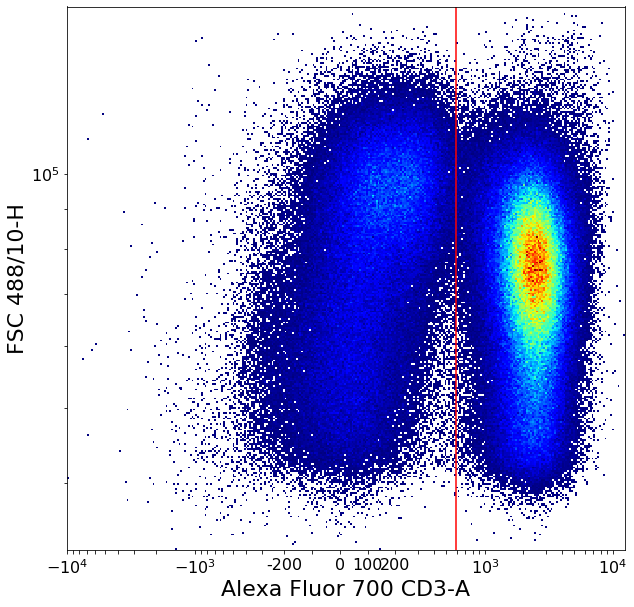
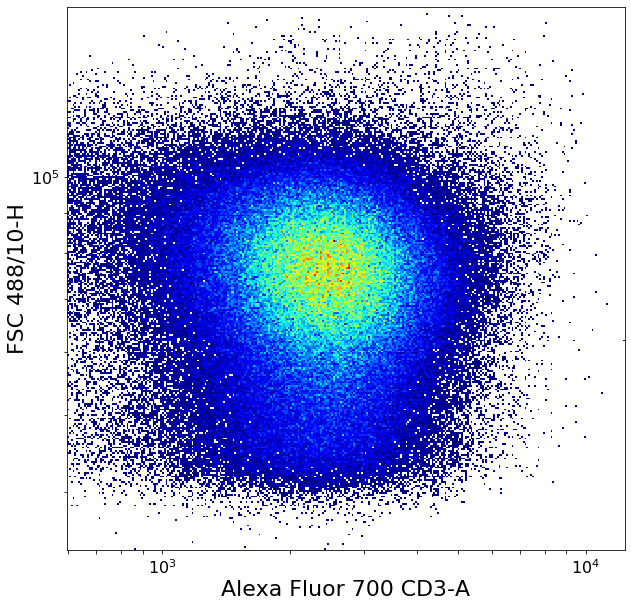
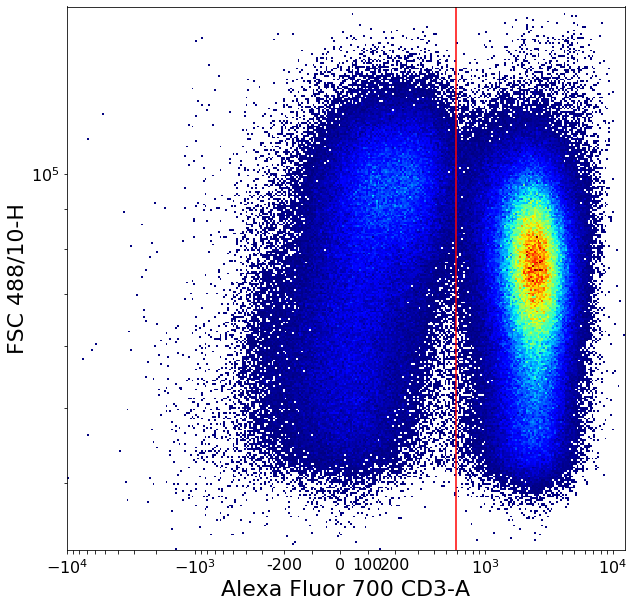
Separating CD3 positives using local minima (valleySeek)¶
Simple “ValleySeek” operation, find the lowest density point in the probability density function in some axis.
[8]:
# CD3pos/neg
xlim = ag.valleySeek(fcs = fcs, xCol = CD3, parentGate = singlets, interval = [0,1000],
require_local_min = True, scale = 'bilog', T = 200)
if xlim == ag.np.inf:
xlim = 400
CD3pos = ag.gateThreshold(fcs = fcs, name = 'CD3pos', xCol = CD3, yCol = "FSC 488/10-H",
parentGate = singlets, thresh = xlim, population = 'upper', scale = 'bilog', T = 200)
CD3neg = ag.gateThreshold(fcs = fcs, name = 'CD3pos', xCol = CD3, yCol = "FSC 488/10-H",
parentGate = singlets, thresh = xlim, population = 'lower', scale = 'bilog', T = 200)
<Figure size 720x720 with 0 Axes>
<Figure size 720x720 with 0 Axes>
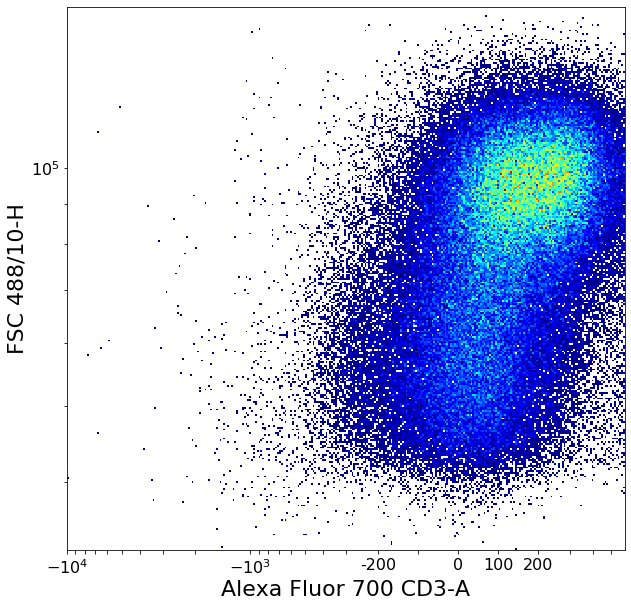
Separating CD4 / CD8 using a quad gate¶
Two valleyseeks like above, then calling QuadGate with the collected limits
[9]:
# CD4/CD8
xlim = ag.valleySeek(fcs, xCol = CD8, parentGate = CD3pos, interval = [0,3000],
require_local_min = True, scale = 'bilog', T = 200)
if xlim == ag.np.inf:
xlim = 1500
CD8neg_tmp = ag.gateThreshold(fcs = fcs, name = 'CD8neg_temp', xCol = CD8,
parentGate = CD3pos, thresh = xlim, population = 'lower',
scale = 'bilog', T = 200)
ylim = ag.valleySeek(fcs, xCol = CD4, parentGate = CD8neg_tmp, interval = [50,1000],
require_local_min = True, scale = 'bilog', T = 200)
if ylim == ag.np.inf:
ylim = 200
if ylim < 100: # Additional check for too low values
ylim = 200 # In such cases, set to ~fmo
CD4pos, CD4CD8_doublepos, CD8pos, CD4neg = ag.quadGate(fcs, names = ["CD4pos","CD4CD8_doublepos", "CD8pos", "CD4neg"],
xCol = CD8, yCol = CD4, parentGate = CD3pos, xThresh = xlim,
yThresh = ylim, scale = 'bilog', T = 200)
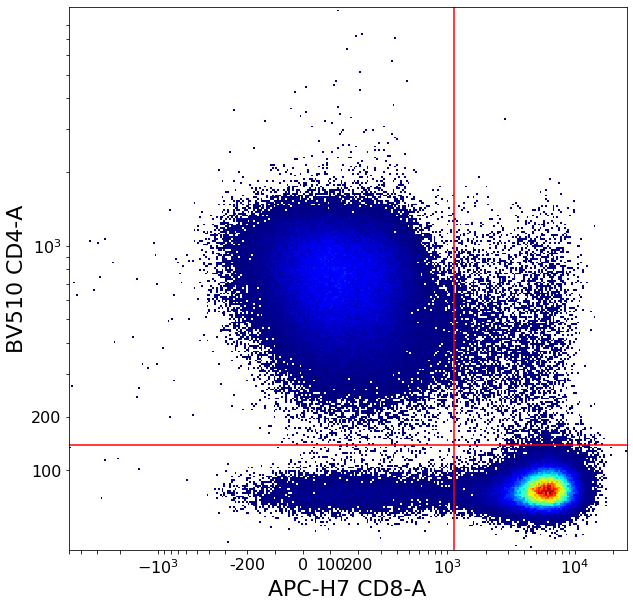
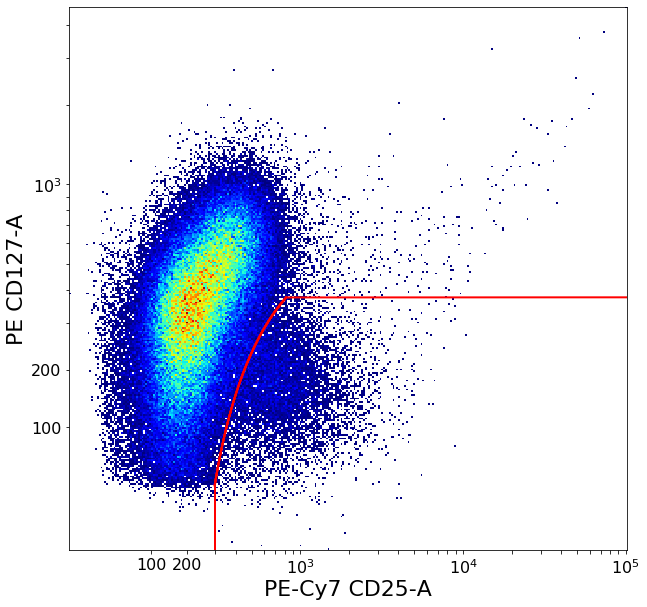
Gating T-reg cells using density delimitation and bezier curves¶
We cut the cluster in strategic sections and try to figure out where it ends in both x- and y-axis using densityDelimitation.
Using these thresholds we then use gateBezier, this gate works similarly to gateCorner but has a smooth transition on it’s edge defined by a bezier curve. We pick one of three preset bezier parameter options based on if the bottom-right part of the main cluster seems to bulge inwards, outwards or is roughly linear.
[10]:
CD4pos_hiCD25 = ag.gateThreshold(fcs, name = "tmp", xCol = CD25,
parentGate = CD4pos, thresh = -200, population = 'upper')
mean, median, sigma, maxVal = ag.axisStats(fcsDF = fcs(), xCol = CD25,
vI = CD4pos_hiCD25(), scale = 'bilog', T = 200)
lim_upper = ag.inverseTransformWrapper(maxVal+0.5*abs(sigma),
scale = 'bilog', T = 200)
lim_lower = ag.inverseTransformWrapper(maxVal-0.5*abs(sigma),
scale = 'bilog', T = 200)
xmaxVal = lim_upper
tmp_maxCD25 = ag.gateThreshold(fcs, name = "tmp", xCol = CD25, parentGate = CD4pos,
thresh = lim_upper, population = 'lower')
maxCD25 = ag.gateThreshold(fcs, name = "tmp", xCol = CD25, parentGate = tmp_maxCD25,
thresh = lim_lower, population='upper')
ylim = ag.densityDelimitation(fcs = fcs, xCol = CD127, parentGate = maxCD25, interval = [-200,1000],
limit_threshold = 0.05, direction = 'left', scale = 'bilog', T = 200)
mean, median, sigma, maxVal = ag.axisStats(fcsDF = fcs(), xCol = CD127,
vI = CD4pos(), scale = 'bilog', T = 200)
lim_upper = ag.inverseTransformWrapper(maxVal+0.5*abs(sigma), scale = 'bilog', T = 200)
lim_lower = ag.inverseTransformWrapper(maxVal-0.5*abs(sigma), scale = 'bilog', T = 200)
ymaxVal = ag.inverseTransformWrapper(maxVal, scale = 'bilog', T = 200)
tmp_maxCD127 = ag.gateThreshold(fcs, name = "tmp", xCol = CD127, parentGate = CD4pos,
thresh = lim_upper, population = 'lower')
maxCD127 = ag.gateThreshold(fcs, name = "tmp", xCol = CD127, parentGate = tmp_maxCD127,
thresh = lim_lower, population = 'upper')
xlim = ag.densityDelimitation(fcs = fcs, xCol = CD25, parentGate = maxCD127, interval = [200,10000],
limit_threshold = 0.05, direction = 'right', scale = 'bilog', T = 200)
if ymaxVal < 0:
ymaxVal = 0
if ymaxVal > 700:
ymaxVal = 700
bezierXParam = [0.7]
bezierYParam = [0.3]
# Take midpoint between ylim and ymaxVal (ymaxval >> ylim) in transformed coordinates
# Midpoint in transformed coordinates (!)
tyMid = (ag.transformWrapper([ymaxVal], T = 200, scale = 'bilog')[0] + ag.transformWrapper([ylim], T = 200, scale = 'bilog')[0]) / 2
# Copy code from above to make a 'street' and calc density delim (we have already calculated the mean and standard deviation)
lim_upper = ag.inverseTransformWrapper(tyMid+0.5*abs(sigma), scale = 'bilog', T = 200)
lim_lower = ag.inverseTransformWrapper(tyMid-0.5*abs(sigma), scale = 'bilog', T = 200)
tmp_midCD127 = ag.gateThreshold(fcs, name = "tmp", xCol = CD127, parentGate = CD4pos,
thresh = lim_upper, population = 'lower')
midCD127 = ag.gateThreshold(fcs, name = "tmp", xCol = CD127, parentGate = tmp_midCD127,
thresh = lim_lower, population = 'upper')
xMid = ag.densityDelimitation(fcs = fcs, xCol = CD25, parentGate = midCD127,
interval = [-200,10000], limit_threshold = 0.3,
direction = 'right', scale = 'bilog', T = 200)
# See which percent this xMid is at from relative to xlim and xmaxVal (xlim >> xmaxVal)
fraction_traveled_x = (xMid-xmaxVal) / (xlim-xmaxVal)
if fraction_traveled_x < 0.30:
# Curve inward
bezierXParam = [0.3]
bezierYParam = [0.7]
elif fraction_traveled_x > 0.70:
# curve outward
bezierXParam = [0.7]
bezierYParam = [0.3]
else:
# straight
bezierXParam = [0.5]
bezierYParam = [0.5]
Tregs = ag.gateBezier(fcs, name = "tmp", xCol = CD25, yCol = CD127, endExtension = 'right',
startExtension = 'down', population = 'lower', parentGate = CD4pos,
points = [(xmaxVal,ylim), (xlim,ymaxVal)], xParam = bezierXParam,
yParam = bezierYParam, scale = 'bilog', T = 200)
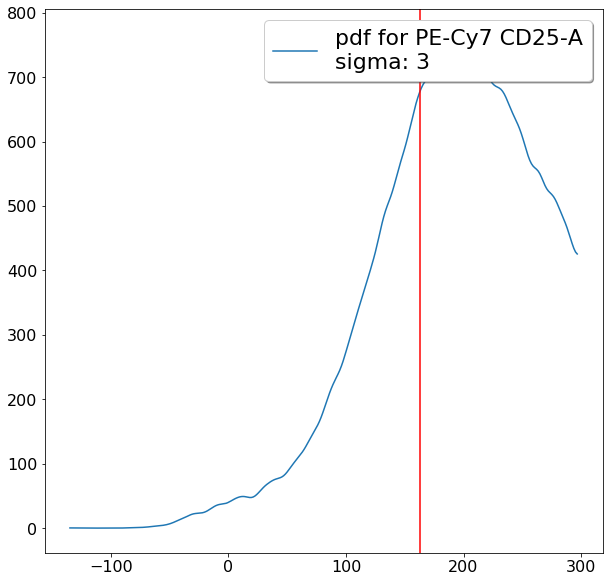
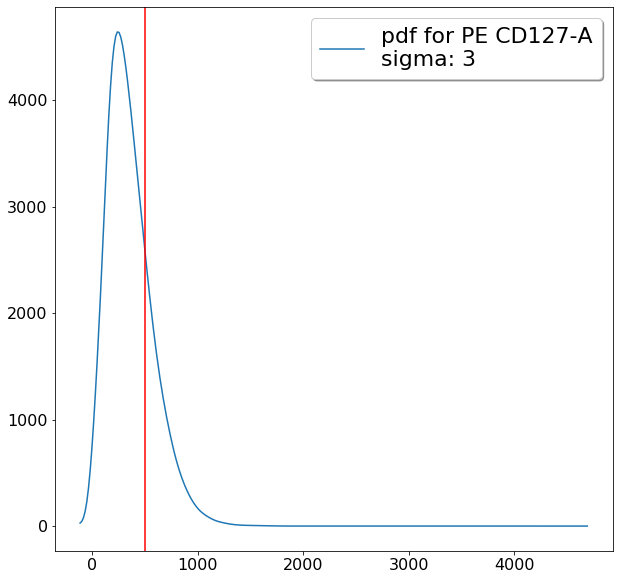
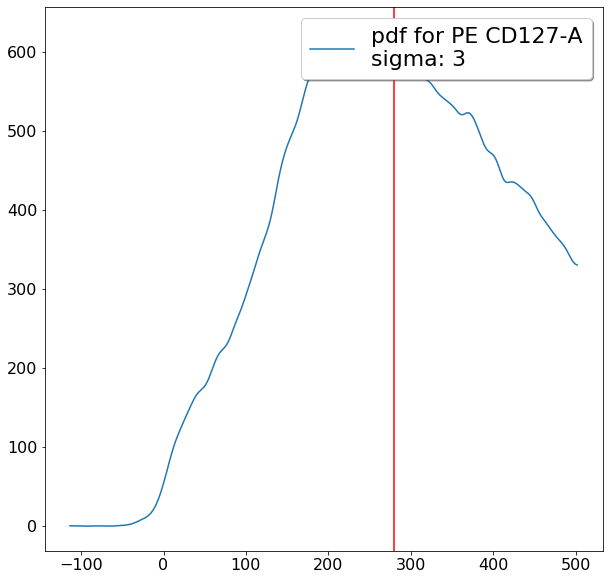
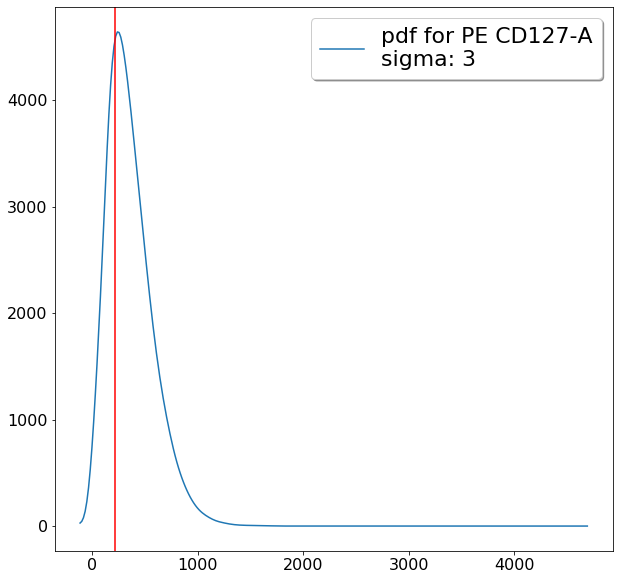
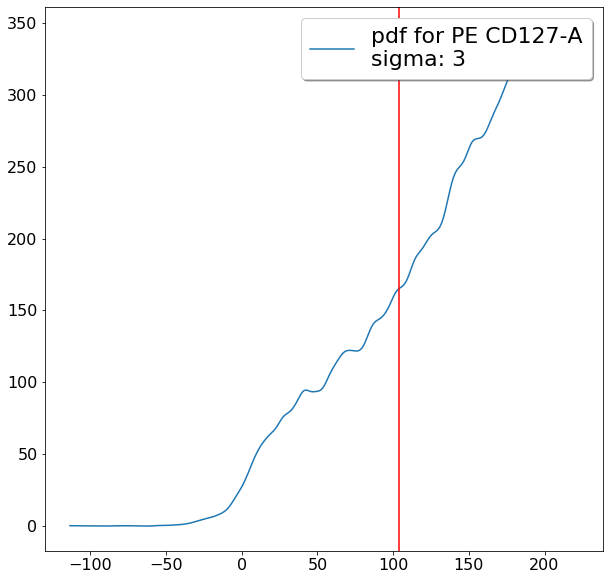
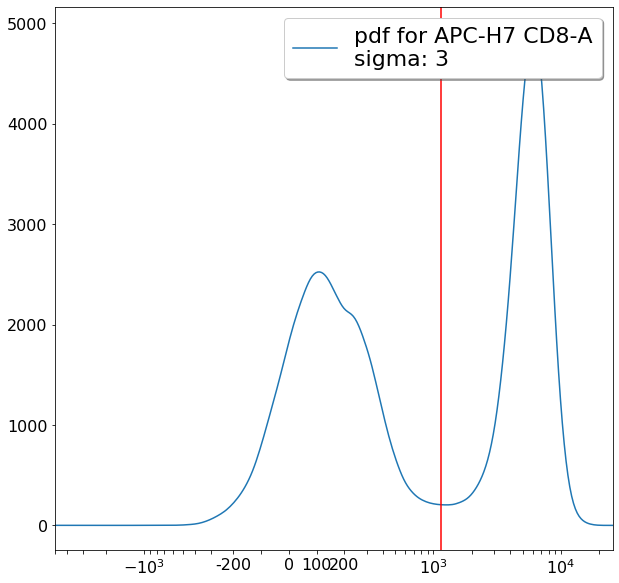
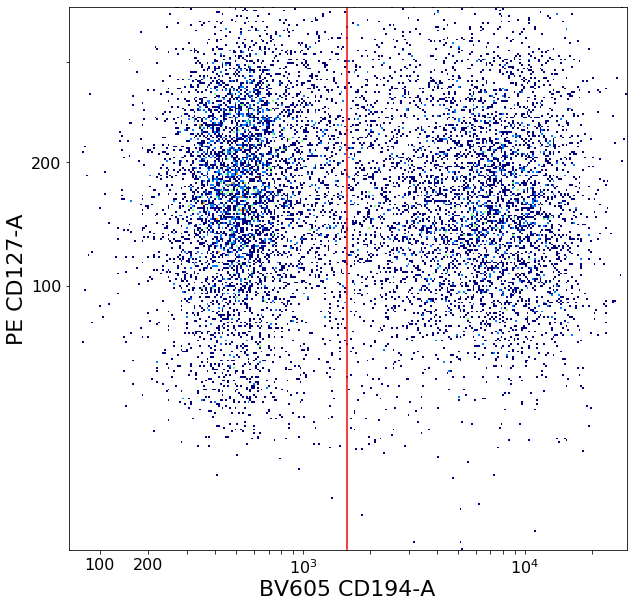
gating CD194 positives (valleySeek)¶
[11]:
# CD194+ T-regs
xlim = ag.valleySeek(fcs, xCol = CD194, parentGate = Tregs, interval = [500,5000], require_local_min = True,
sigma = 3, scale = 'bilog', T = 200)
if xlim == ag.np.inf:
xlim = 1500
CD194_Tregs = ag.gateThreshold(fcs = fcs, name = "CD194_Tregs", xCol = CD194, yCol = CD127,
parentGate = Tregs, thresh = xlim, orientation = 'vertical',
scale = 'bilog', T = 200, population = 'upper')
<Figure size 720x720 with 0 Axes>
[12]:
# Tregs subpopulations - activated, secreting, resting
# First gate out the resting (top) by estimating half-normal distribution on bottom cluster.
# Then look at the CD25 expression of these resting Tregs, and set the limit between secreting and activated Tregs
# at some point of the right-tail of the CD25 distribution
# First do arbitrary cut in the y-axis somewhere between 0-1000
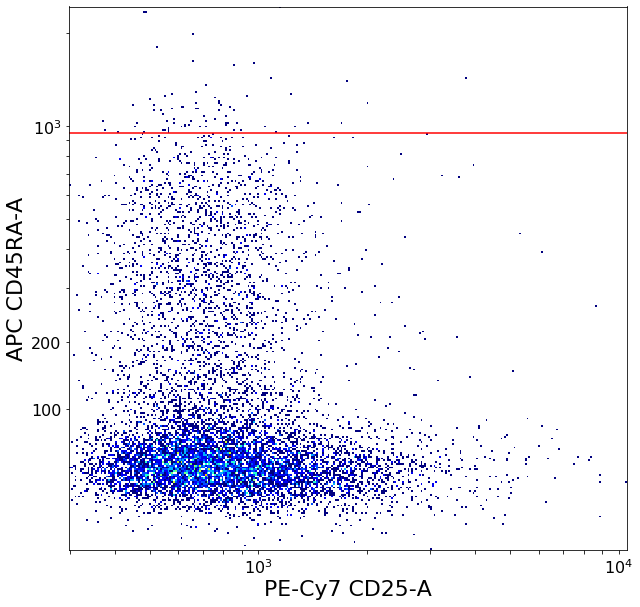
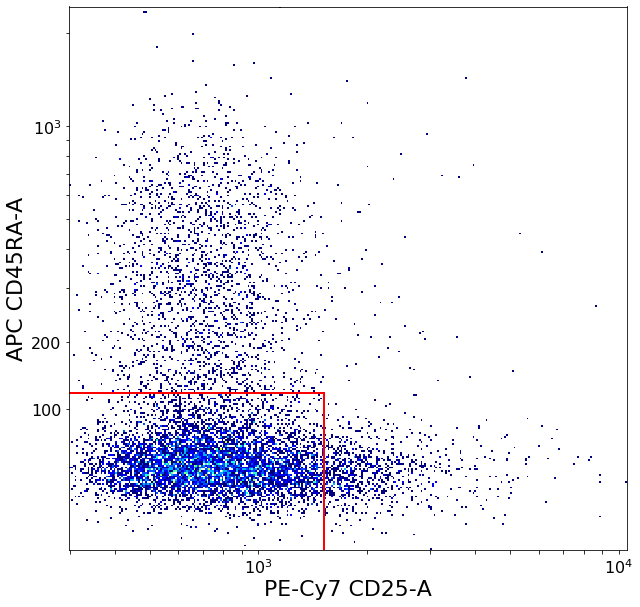
ylim = ag.valleySeek(fcs, xCol = CD45RA, parentGate = Tregs,
interval = [0,1000], scale = 'bilog', T = 200)
tmpbottomTreg = ag.gateThreshold(fcs, xCol = CD25, yCol = CD45RA, name = "bottomTreg",
parentGate = Tregs, thresh = ylim, population='lower',
orientation = 'horizontal', scale = 'bilog', T = 200, update = False)
# In this population, find the most dense point
tmp, tmp1, tmp2, maxVal = ag.axisStats(fcs(), xCol = CD45RA, vI = tmpbottomTreg(), scale = 'bilog', T = 200)
maxVal = ag.inverseTransformWrapper(maxVal, scale = 'bilog', T = 200)
mean, sigma = ag.halfNormalDistribution(fcs, xCol = CD45RA, mean = maxVal, direction = 'left', parentGate = tmpbottomTreg,
scale = 'bilog', T = 200)
# Now we can set the final ylim with big margin (4 std devs):
ylim = ag.inverseTransformWrapper(mean + 4*abs(sigma), scale = 'bilog', T = 200)
if ylim > 1000:
ylim = 1000
if ylim < 100:
ylim = 100
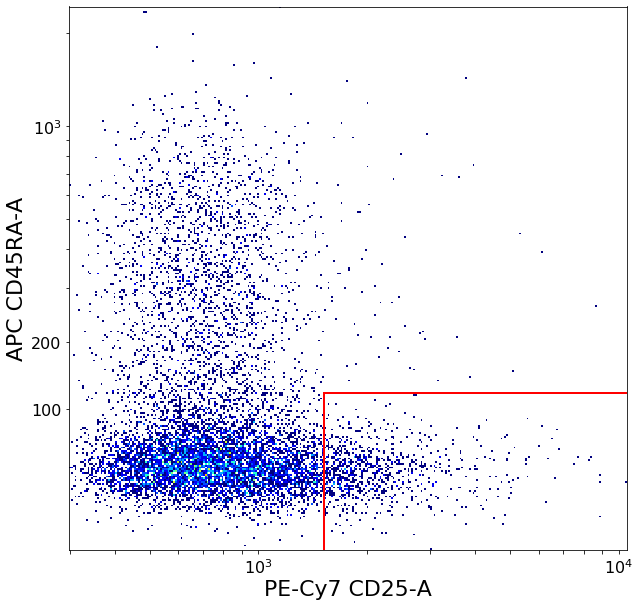
# We use this to gate the activated Tregs
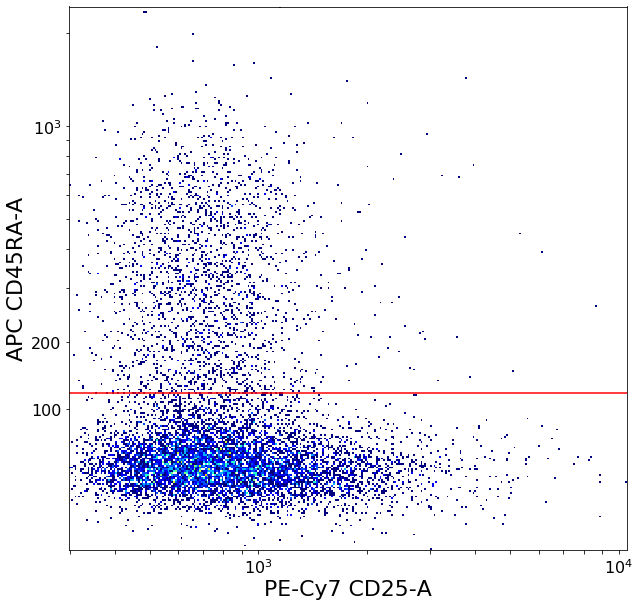
restingTreg = ag.gateThreshold(fcs, xCol = CD25, yCol = CD45RA, name = "restingTreg", thresh = ylim, parentGate = Tregs,
scale = 'bilog', T = 200, population = 'upper', orientation = 'horizontal', update = False, filePlot = None)
# We then investigate the CD25 distribution in this population and set the treshold at one standard dev on the right tail
mean, median, sigma, maxVal = ag.axisStats(fcs(), xCol = CD25, vI = restingTreg(), scale = 'bilog', T = 200)
# With this we can set the final xlim
xlim = mean + 2*sigma
xlim = ag.inverseTransformWrapper(xlim, scale = 'bilog', T = 200)
if xlim > 10000:
xlim = 10000
if xlim < 1000:
xlim = 1000
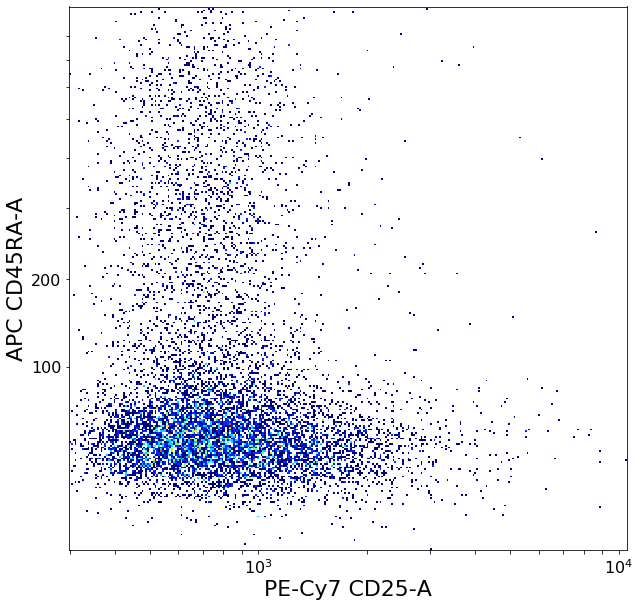
activatedTreg = ag.gateCorner(fcs, name = "activatedTreg", xCol = CD25, yCol = CD45RA, xThresh = xlim, yThresh = ylim,
xOrientation = "upper", yOrientation = "lower", parentGate = Tregs, scale = 'bilog', T = 200, filePlot = None)
secretingTreg = ag.gateCorner(fcs, name = "secretingTreg", xCol = CD25, yCol = CD45RA, xThresh = xlim, yThresh = ylim, xOrientation = "lower",
yOrientation = "lower", parentGate = Tregs, scale = 'bilog', T = 200, filePlot = None)
<Figure size 720x720 with 0 Axes>
<Figure size 720x720 with 0 Axes>
T-helper subsets (valleySeek and densityDelimitation)¶
Th1, Th2, Th17
Simple valleyseek to separate the two main clusters, then estimate where the rough edge of the cluster in the CXCR3 axis using densityDelimitation.
Using these thresholds, a call to quadGate can separate the populations
[13]:
follicular_xlim = ag.valleySeek(fcs, xCol = CCR6, interval = [0,1000], require_local_min = True, parentGate = CD4pos,
scale = 'bilog', T = 200, sigma = 2)
follicular_ylim = ag.densityDelimitation(fcs, xCol = CXCR3, parentGate = CD4pos, limit_threshold = 0.2,
scale = 'bilog', T = 200, direction = 'right', interval = [100,1000], sigma = 1)
Th1, Th2, Th1_Th17, Th17 = ag.quadGate(fcs = fcs, names = ['Th1', 'Th2', 'Th1_Th17', 'Th17'], xCol = CCR6, yCol = CXCR3,
xThresh = follicular_xlim, yThresh = follicular_ylim, parentGate = CD4pos, scale = 'bilog', T = 200)
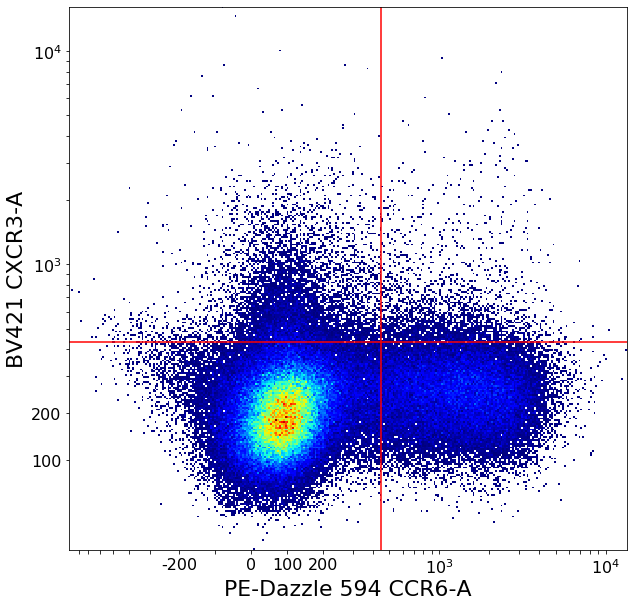
CXCR5 positives (valleySeek and densityDelimitation)¶
Very similar strategy as for the subsets above
[14]:
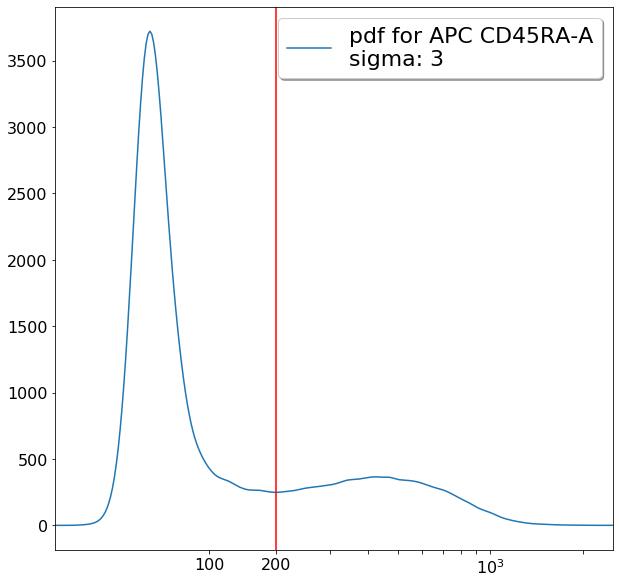
# Parent is still CD4pos
xlim = ag.valleySeek(fcs, xCol = CD45RA, parentGate = CD4pos, interval = [0,700],
require_local_min = True, sigma = 3, scale = 'bilog', T = 200)
if xlim == ag.np.inf:
xlim = 200
if xlim < 200:
xlim = 200
tmpHiCD45RA = ag.gateThreshold(fcs, xCol = CD45RA, name = "tmpHiCD45RA", parentGate = CD4pos,
thresh = xlim, population = 'upper', orientation = 'vertical',
scale = 'bilog', T = 200, update = False)
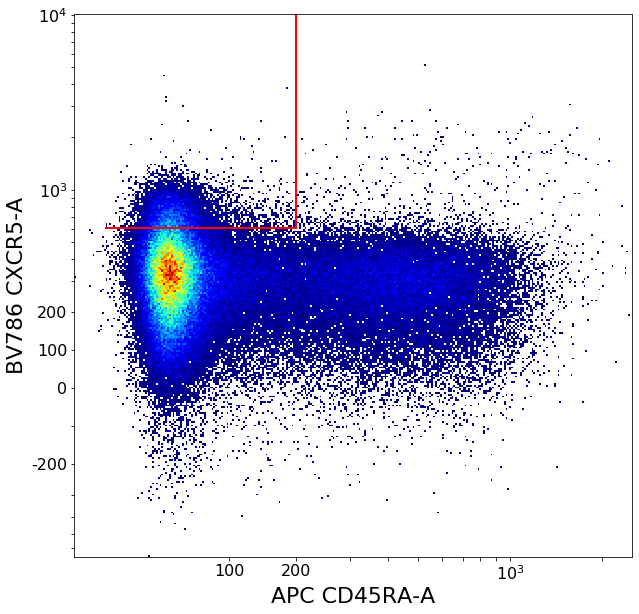
ylim = ag.densityDelimitation(fcs, xCol = CXCR5, parentGate = tmpHiCD45RA, interval = [0,2500],
direction = 'right', limit_threshold = 0.1, scale = 'bilog', T = 200)
CXCR5pos = ag.gateCorner(fcs, name = 'CXCR5pos', xCol = CD45RA, yCol = CXCR5, parentGate = CD4pos,
xThresh = xlim, yThresh = ylim, xOrientation = 'lower', yOrientation = 'upper',
scale = 'bilog', T = 200)
Follicular T-helper cells (densityDelimitation)¶
This gate’s purpose is mainly to remove some scattered unspecific events and get a purer Th cluster.
Two calls to densityDelimitation to find the edges of the main cluster in both directions, then a fixed bezierGate to roughly cut out unspecific events.
[15]:
xlim = ag.densityDelimitation(fcs, xCol = CD25, parentGate = CXCR5pos, interval = [0,3000], direction = 'right',
limit_threshold = 0.05, scale = 'bilog', T = 200)
if xlim == ag.np.inf:
xlim = 500
ylim = ag.densityDelimitation(fcs, xCol = CD127, parentGate = CXCR5pos, interval = [0,2000], direction = 'left',
limit_threshold = 0.1, scale = 'bilog', T = 200)
if ylim == ag.np.inf:
ylim = 300
Follicular_Th = ag.gateBezier(fcs, name = 'some_TH', xCol = CD25, yCol = CD127, parentGate = CXCR5pos,
endExtension = 'up', startExtension = 'left', points = [(300,ylim), (xlim,1000)],
population = 'upper', xParam = [0.8], yParam = [0.8], scale = 'bilog', T = 200)
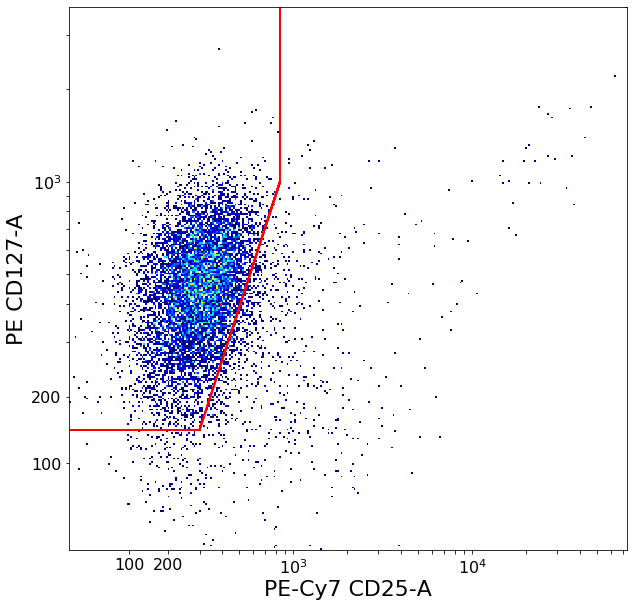
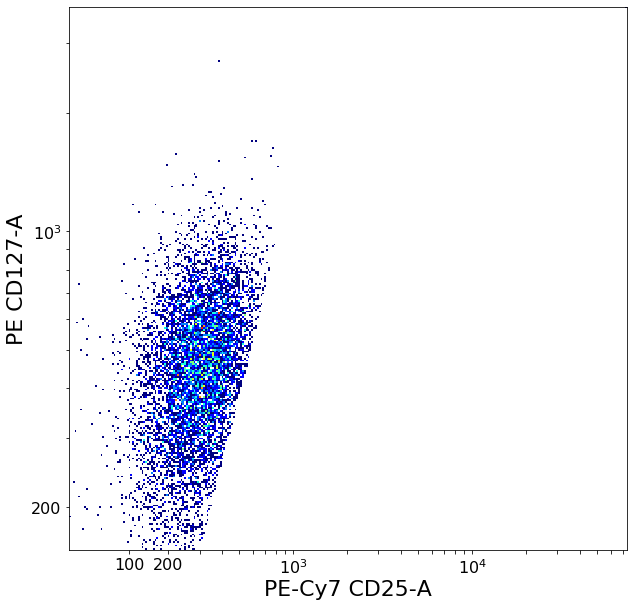
Follicular T-helper subsets (quadGate)¶
This gate uses the thresholds defined a few gates up for normal T-helper subsets, but now applied to follicular T-helpers
[16]:
# This gate recycles the thresholds from the TH1,TH17, GATE (two steps up!)
# Since this gate tries to do the same separation but on a smaller subset of events
Follicular_th1, Follicular_th1_th17, Follicular_th17, Follicular_Th2 = ag.quadGate(fcs = fcs,
names=['Follicular_th1', 'Follicular_th1-th17', 'Follicular_th17', 'Follicular_Th2'],
xCol = CCR6, yCol = CXCR3, xThresh = follicular_xlim, yThresh = follicular_ylim,
parentGate = Follicular_Th, scale = 'bilog', T = 200)
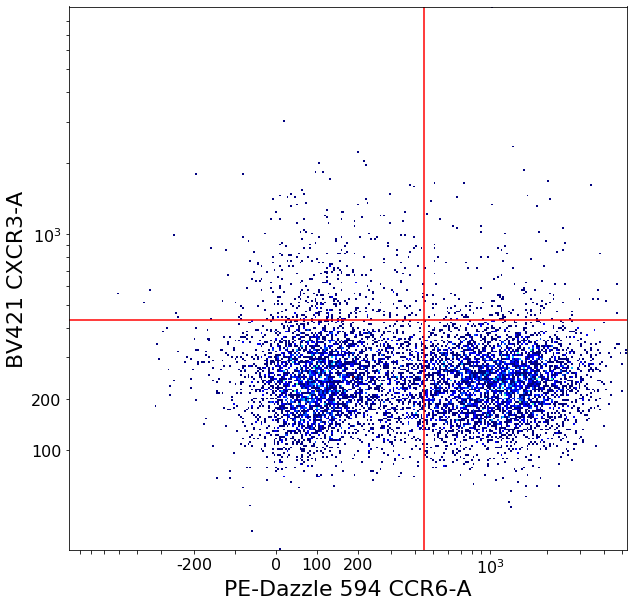
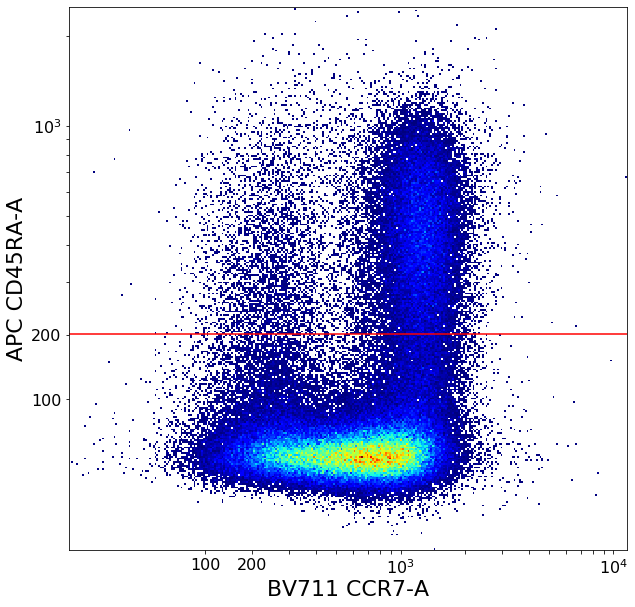
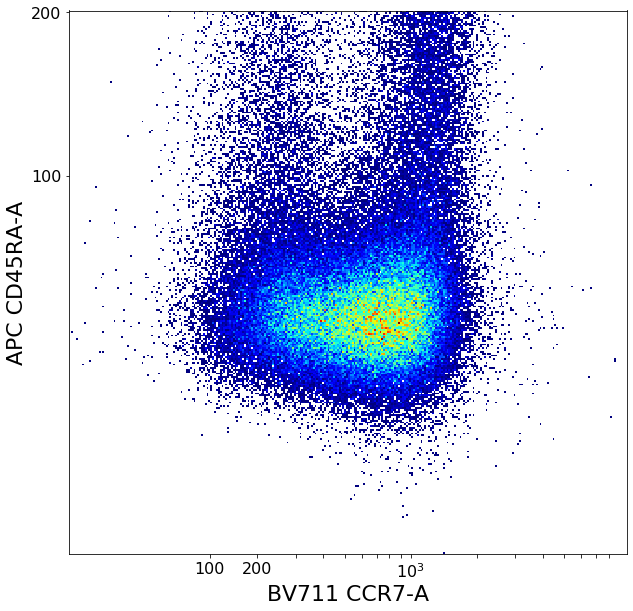
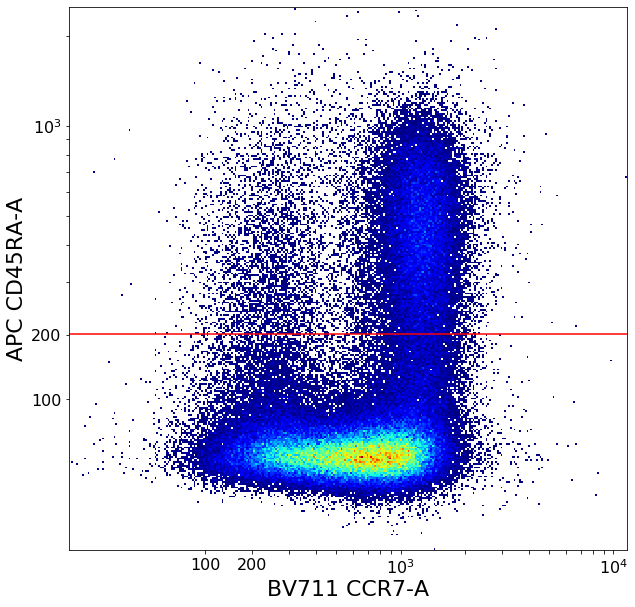
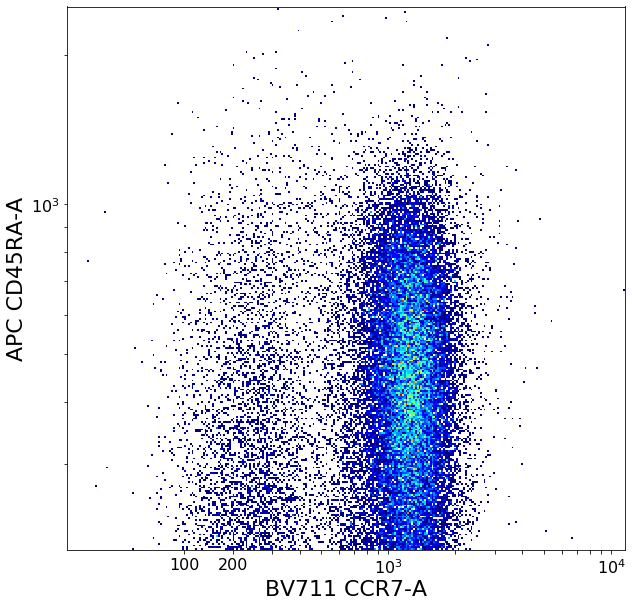
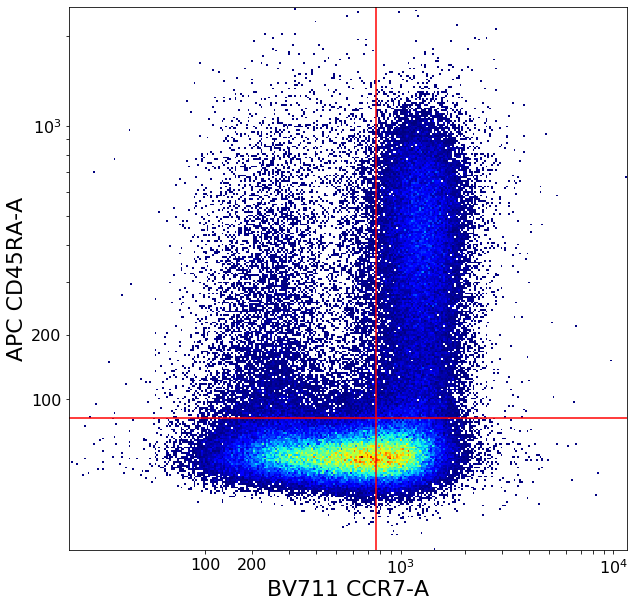
CD4 positive subsets (valleySeek and quadGate)¶
First tries to find a top-bottom separation with valleySeek, and branches into two options depending on if it finds one or not.
Using this separation the algorithm tries to estimate the height of the bottom cluster, and the width of the top cluster using densityDelimitation. Using these two threshold, a quadGate is called.
[17]:
# Find initial top-bottom rough separation
ylim = ag.valleySeek(fcs, xCol = CD45RA, interval = [0,800], require_local_min = True, parentGate = CD4pos, scale = 'bilog', T = 200)
if ylim == ag.np.inf:
ylim = 200
bNoYlimSep = True
else:
bNoYlimSep = False
if ylim < 200:
ylim=200
tempCD4quad_bottom = ag.gateThreshold(fcs, xCol = CCR7, yCol = CD45RA,name = "bottomTmp", parentGate = CD4pos, thresh = ylim,
population = 'lower', orientation = 'horizontal', scale = 'bilog', T = 200,update = False)
tempCD4quad_top = ag.gateThreshold(fcs, xCol = CCR7, yCol = CD45RA, name = "topTmp", parentGate = CD4pos, thresh = ylim,
population = 'upper', orientation = 'horizontal', scale = 'bilog', T = 200,update = False)
# Fine adjust limits based on those two clusters
# If no proper separation of clusters - increase the density delim
if bNoYlimSep:
ylim = ag.densityDelimitation(fcs, xCol = CD45RA, interval = [0,1000], direction = 'right', limit_threshold = 0.5,
parentGate = tempCD4quad_bottom, scale = 'bilog', T = 200)
else:
ylim = ag.densityDelimitation(fcs, xCol = CD45RA, interval = [0,1000], direction = 'right', limit_threshold = 0.2,
parentGate = tempCD4quad_bottom, scale = 'bilog', T = 200)
if ylim == ag.np.inf:
ylim = 200
xlim = ag.densityDelimitation(fcs, xCol = CCR7, interval = [500,2000], direction = 'left', limit_threshold = 0.2,
parentGate = tempCD4quad_top, scale = 'bilog', T = 200)
if xlim == ag.np.inf:
xlim = 600
# Gate
effector_CD4pos, naive_CD4pos, central_memory_CD4pos, effector_memory_CD4pos = ag.quadGate(fcs = fcs, names = ['effector_CD4pos', 'naive_CD4pos', 'central_memory_CD4pos', 'effector_memory_CD4pos'],
xCol = CCR7, yCol = CD45RA, xThresh = xlim, yThresh = ylim, parentGate = CD4pos, scale = 'bilog', T = 200)
<Figure size 720x720 with 0 Axes>
<Figure size 720x720 with 0 Axes>
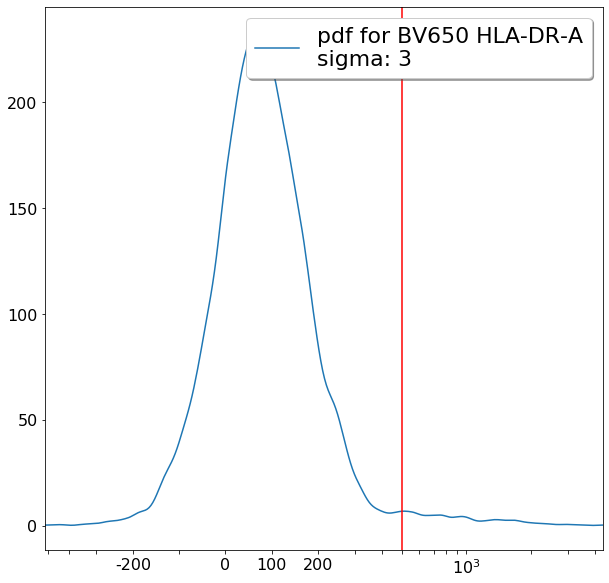
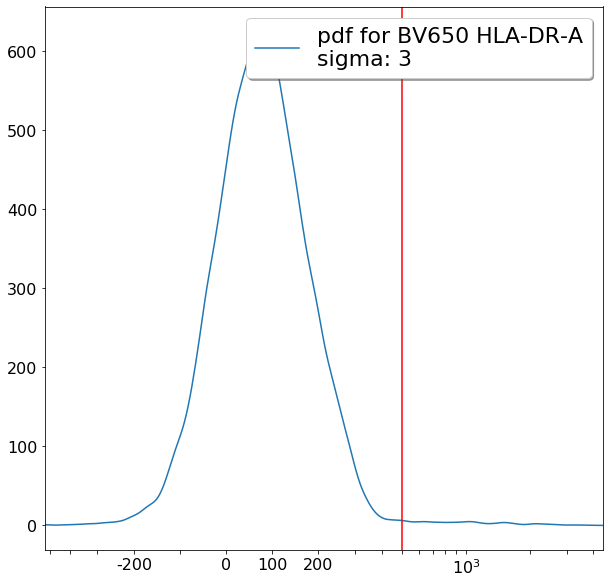
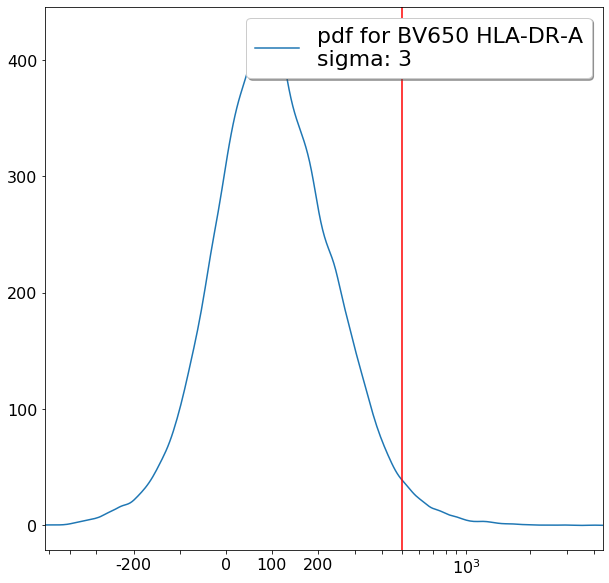
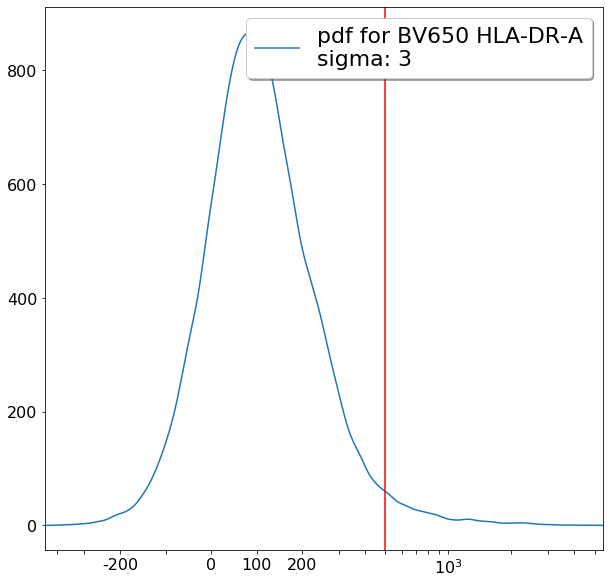
FMO based thresholds for HLA-DR positivity (gateThreshold)¶
[18]:
effector_CD4pos_HLADRpos = ag.gateThreshold(fcs, xCol = HLADR, name = "effector_CD4pos_HLADRpos", parentGate=effector_CD4pos,
thresh = 500, population = 'upper', orientation = 'vertical', scale = 'bilog', T = 200,update = False)
naive_CD4pos_HLADRpos = ag.gateThreshold(fcs, xCol = HLADR, name = "naive_CD4pos_HLADRpos", parentGate=naive_CD4pos,
thresh = 500, population = 'upper', orientation = 'vertical', scale = 'bilog', T = 200, update = False)
central_memory_CD4pos_HLADRpos = ag.gateThreshold(fcs, xCol = HLADR, name = "central_memory_CD4pos_HLADRpos", parentGate=central_memory_CD4pos,
thresh = 500, population = 'upper', orientation = 'vertical', scale = 'bilog', T = 200, update = False)
effector_memory_CD4pos_HLADRpos = ag.gateThreshold(fcs, xCol = HLADR, name = "effector_memory_CD4pos_HLADRpos", parentGate = effector_memory_CD4pos,
thresh = 500, population = 'upper', orientation = 'vertical', scale = 'bilog', T = 200, update = False)
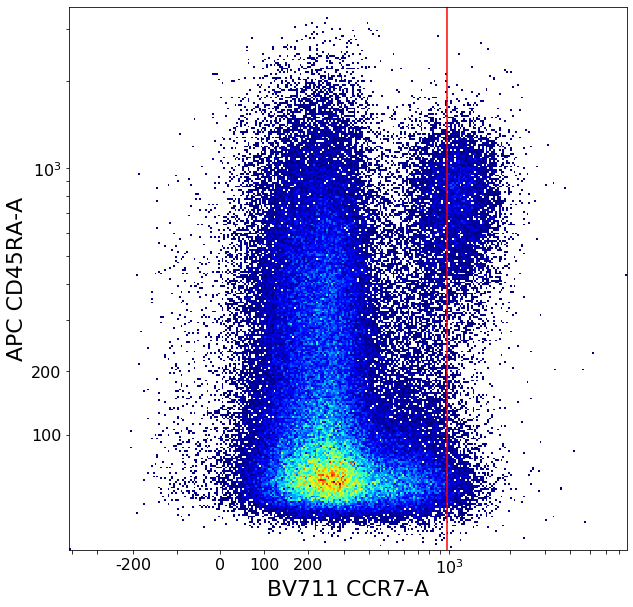
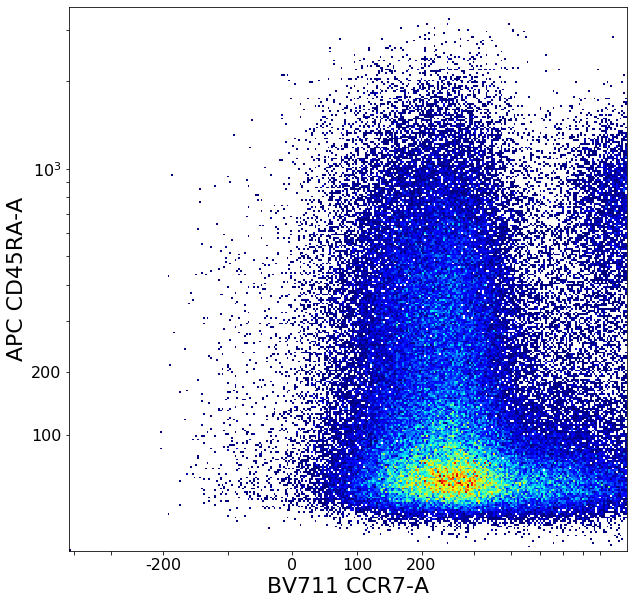
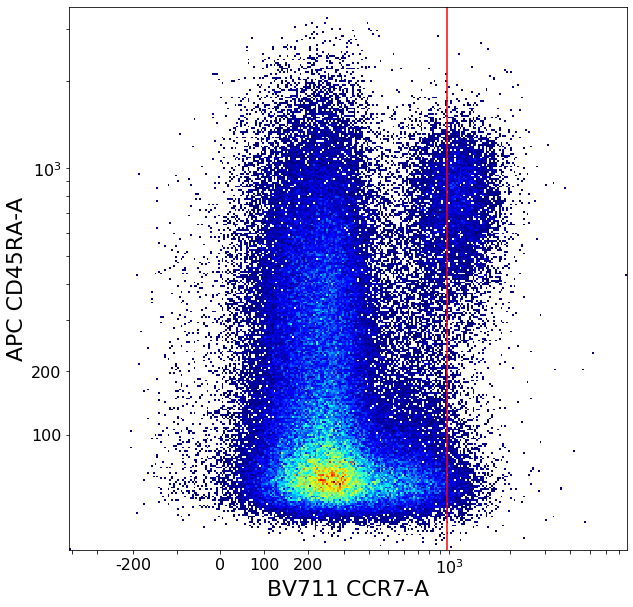
CD8 positive subsets (valleySeek and quadGate)¶
[19]:
# Find initial top-bottom rough separation
xlim = ag.valleySeek(fcs, xCol = CCR7, interval = [200,1500], require_local_min = True, parentGate = CD8pos, scale = 'bilog', T = 200)
if xlim == ag.np.inf:
xlim = 500
if xlim < 500:
xlim = 500
tempCD8quad_left = ag.gateThreshold(fcs, xCol = CCR7, yCol = CD45RA, name = "leftTmp", parentGate = CD8pos, thresh = xlim, population = 'lower',
orientation = 'vertical', scale = 'bilog', T = 200, update = False)
tempCD8quad_right = ag.gateThreshold(fcs, xCol = CCR7, yCol = CD45RA, name = "rightTmp", parentGate = CD8pos, thresh = xlim, population = 'upper',
orientation = 'vertical', scale = 'bilog', T = 200, update = False)
# Fine adjust limits based on those two clusters
ylim = ag.densityDelimitation(fcs, xCol = CD45RA, interval = [0,800], direction = 'right', limit_threshold = 0.3,
parentGate = tempCD8quad_left, scale = 'bilog', T = 200)
if ylim == ag.np.inf:
ylim = 200
# Also check density delimitation using the right half of the cluster
# Reasoning:
# If the left half of the quad is 'smeared' and the right half is quite dense (see sample 9704538)
# Then the ylim based on the right half makes more sense
# Check using that side. If that limit is closer to 200 (the 'default'/fmo) and hasn't hit rock bottom (<100)
# Then use that instead, it's probably better.
# If that side is also smeared, its probably a bad sample...
ylim_alternate = ag.densityDelimitation(fcs, xCol = CD45RA, interval = [0,800], direction = 'left',
limit_threshold = 0.3, parentGate = tempCD8quad_right, scale = 'bilog', T = 200)
if ylim != ag.np.inf:
if abs(ylim_alternate - 200) < abs(ylim - 200) and ylim_alternate > 100:
ylim = ylim_alternate
xlim = ag.densityDelimitation(fcs, xCol = CCR7, interval = [0,1500], direction = 'left', limit_threshold = 0.2,
parentGate = tempCD8quad_right, scale = 'bilog', T = 200)
if xlim == ag.np.inf:
xlim = 500
effector_CD4neg, naive_CD4neg, central_memory_CD4neg, effector_memory_CD4neg = ag.quadGate(fcs = fcs, names = ['effector_CD4neg', 'naive_CD4neg', 'central_memory_CD4neg', 'effector_memory_CD4neg'],
xCol = CCR7, yCol = CD45RA, xThresh = xlim, yThresh = ylim, parentGate = CD8pos, scale = 'bilog', T = 200)
<Figure size 720x720 with 0 Axes>
<Figure size 720x720 with 0 Axes>
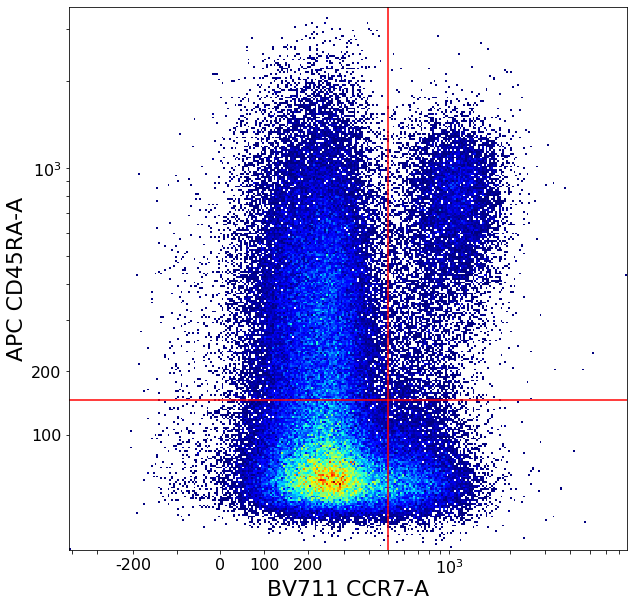
Some more FMO based thresholds for HLA-DR positivity (gateThreshold)¶
[20]:
effector_CD4neg_HLADRpos = ag.gateThreshold(fcs, xCol = HLADR, name = "effector_CD4neg_HLADRpos", parentGate = effector_CD4neg,
thresh = 500, population = 'upper', orientation = 'vertical', scale = 'bilog',
T = 200, update = False)
naive_CD4neg_HLADRpos = ag.gateThreshold(fcs, xCol = HLADR, name = "naive_CD4neg_HLADRpos", parentGate = naive_CD4neg,
thresh = 500, population = 'upper', orientation = 'vertical', scale = 'bilog',
T = 200, update = False)
central_memory_CD4neg_HLADRpos = ag.gateThreshold(fcs, xCol = HLADR, name = "central_memory_CD4neg_HLADRpos", parentGate=central_memory_CD4neg,
thresh = 500, population = 'upper', orientation = 'vertical', scale = 'bilog',
T = 200, update = False)
effector_memory_CD4neg_HLADRpos = ag.gateThreshold(fcs, xCol = HLADR, name = "effector_memory_CD4neg_HLADRpos", parentGate = effector_memory_CD4neg,
thresh = 500, population = 'upper', orientation = 'vertical',
scale = 'bilog', T = 200, update = False)
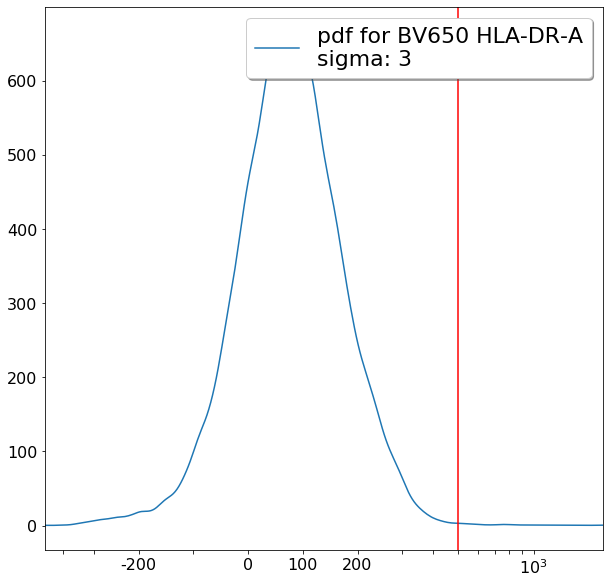
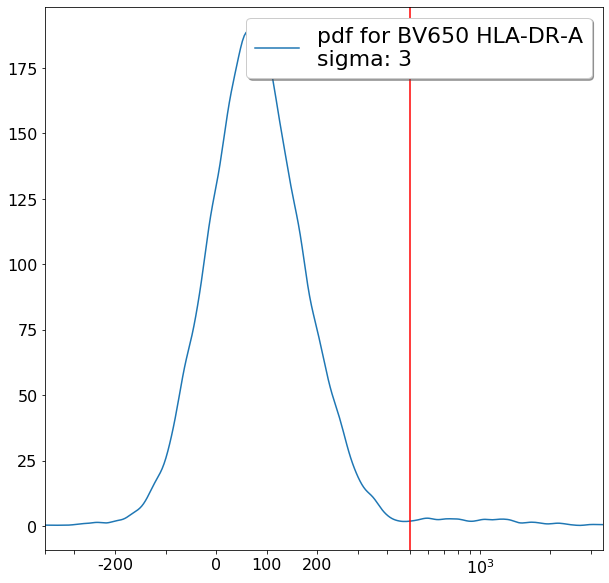
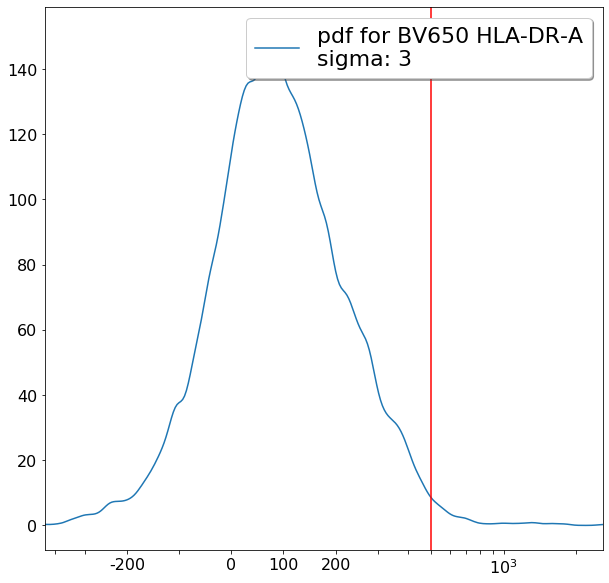
[21]:
# DONE